Timeseries Tutorial
This notebook demonstrates the use of timeseries module capabilities to process data for analysis and build expected energy models.
[20]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import os
import warnings
warnings.filterwarnings("ignore")
[21]:
from pvops.timeseries import preprocess
from pvops.timeseries.models import linear, iec
from pvops.text2time import utils as t2t_utils, preprocess as t2t_preprocess
[22]:
example_prodpath = os.path.join('example_data', 'example_prod_with_covariates.csv')
Load in data
[23]:
prod_data = pd.read_csv(example_prodpath, on_bad_lines='skip', engine='python')
Production data is loaded in with the columns: - date: a time stamp - randid: site ID - generated_kW: generated energy - expected_kW: expected energy - irrad_poa_Wm2: irradiance - temp_amb_C: ambient temperature - wind_speed_ms: wind speed - temp_mod_C: module temperature
[24]:
prod_data.head()
[24]:
| date | randid | generated_kW | expected_kW | irrad_poa_Wm2 | temp_amb_C | wind_speed_ms | temp_mod_C | |
|---|---|---|---|---|---|---|---|---|
| 0 | 2018-04-01 07:00:00 | R15 | 0.475 | 0.527845 | 0.02775 | 16.570 | 4.2065 | 14.1270 |
| 1 | 2018-04-01 08:00:00 | R15 | 1332.547 | 1685.979445 | 87.91450 | 16.998 | 4.1065 | 15.8610 |
| 2 | 2018-04-01 09:00:00 | R15 | 6616.573 | 7343.981135 | 367.90350 | 20.168 | 4.5095 | 24.5745 |
| 3 | 2018-04-01 10:00:00 | R15 | 8847.800 | 10429.876422 | 508.28700 | 21.987 | 4.9785 | 30.7740 |
| 4 | 2018-04-01 11:00:00 | R15 | 11607.389 | 12981.228814 | 618.79450 | 23.417 | 4.6410 | 35.8695 |
[25]:
metadata = pd.DataFrame()
metadata['randid'] = ['R15', 'R10']
metadata['dcsize'] = [20000, 20000]
metadata.head()
[25]:
| randid | dcsize | |
|---|---|---|
| 0 | R15 | 20000 |
| 1 | R10 | 20000 |
Column dictionaries
Create production and metadata column dictionary with format {pvops variable: user-specific column names}. This establishes a connection between the user’s data columns and the pvops library.
[26]:
prod_col_dict = {'siteid': 'randid',
'timestamp': 'date',
'powerprod': 'generated_kW',
'irradiance':'irrad_poa_Wm2',
'temperature':'temp_amb_C', # Optional parameter, used by one of the modeling structures
'baseline': 'IEC_pstep', #user's name choice for new column (baseline expected energy defined by user or calculated based on IEC)
'dcsize': 'dcsize', #user's name choice for new column (System DC-size, extracted from meta-data)
'compared': 'Compared',#user's name choice for new column
'energy_pstep': 'Energy_pstep', #user's name choice for new column
'capacity_normalized_power': 'capacity_normalized_power', #user's name choice for new column
}
metad_col_dict = {'siteid': 'randid',
'dcsize': 'dcsize'}
Data Formatting
Use the prod_date_convert function to convert date information to python datetime objects and use prod_nadate_process to handle data entries with no date information - here we use pnadrop=True to drop such entries.
[27]:
prod_data_converted = t2t_preprocess.prod_date_convert(prod_data, prod_col_dict)
prod_data_datena_d, _ = t2t_preprocess.prod_nadate_process(prod_data_converted, prod_col_dict, pnadrop=True)
Assign production data index to timestamp data, using column dictionary to translate to user columns.
[28]:
prod_data_datena_d.index = prod_data_datena_d[prod_col_dict['timestamp']]
min(prod_data_datena_d.index), max(prod_data_datena_d.index)
[28]:
(Timestamp('2018-04-01 07:00:00'), Timestamp('2019-03-31 18:00:00'))
[29]:
prod_data_datena_d
[29]:
| date | randid | generated_kW | expected_kW | irrad_poa_Wm2 | temp_amb_C | wind_speed_ms | temp_mod_C | |
|---|---|---|---|---|---|---|---|---|
| date | ||||||||
| 2018-04-01 07:00:00 | 2018-04-01 07:00:00 | R15 | 0.475 | 0.527845 | 0.02775 | 16.570000 | 4.2065 | 14.127000 |
| 2018-04-01 08:00:00 | 2018-04-01 08:00:00 | R15 | 1332.547 | 1685.979445 | 87.91450 | 16.998000 | 4.1065 | 15.861000 |
| 2018-04-01 09:00:00 | 2018-04-01 09:00:00 | R15 | 6616.573 | 7343.981135 | 367.90350 | 20.168000 | 4.5095 | 24.574500 |
| 2018-04-01 10:00:00 | 2018-04-01 10:00:00 | R15 | 8847.800 | 10429.876422 | 508.28700 | 21.987000 | 4.9785 | 30.774000 |
| 2018-04-01 11:00:00 | 2018-04-01 11:00:00 | R15 | 11607.389 | 12981.228814 | 618.79450 | 23.417000 | 4.6410 | 35.869500 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2019-03-31 14:00:00 | 2019-03-31 14:00:00 | R10 | 19616.000 | 20179.797303 | 967.10350 | -4.129722 | 5.9935 | 35.860694 |
| 2019-03-31 15:00:00 | 2019-03-31 15:00:00 | R10 | 19552.000 | 20149.254116 | 983.94500 | -4.214444 | 5.1770 | 38.549306 |
| 2019-03-31 16:00:00 | 2019-03-31 16:00:00 | R10 | 19520.000 | 20192.672439 | 1011.77950 | -3.943611 | 4.2430 | 42.848889 |
| 2019-03-31 17:00:00 | 2019-03-31 17:00:00 | R10 | 17968.000 | 19141.761319 | 895.76800 | -4.124167 | 3.1885 | 42.321806 |
| 2019-03-31 18:00:00 | 2019-03-31 18:00:00 | R10 | 11616.000 | 12501.159607 | 649.48700 | -4.663056 | 2.2540 | 38.596944 |
8755 rows × 8 columns
Data Preprocessing
Preprocess data with prod_inverter_clipping_filter using the threshold model. This adds a mask column to the dataframe where True indicates a row to be removed by the filter.
[30]:
masked_prod_data = preprocess.prod_inverter_clipping_filter(prod_data_datena_d, prod_col_dict, metadata, metad_col_dict, 'threshold', freq=60)
filtered_prod_data = masked_prod_data[masked_prod_data['mask'] == False]
print(f"Detected and removed {sum(masked_prod_data['mask'])} rows with inverter clipping.")
Detected and removed 24 rows with inverter clipping.
Normalize the power by capacity
[31]:
for site in metadata[metad_col_dict['siteid']].unique():
site_metad_mask = metadata[metad_col_dict['siteid']] == site
site_prod_mask = filtered_prod_data[prod_col_dict['siteid']] == site
dcsize = metadata.loc[site_metad_mask, metad_col_dict['dcsize']].values[0]
filtered_prod_data.loc[site_prod_mask, [prod_col_dict['capacity_normalized_power']]] = filtered_prod_data.loc[site_prod_mask, prod_col_dict['powerprod']] / dcsize
Visualize the power signal versus covariates for one site with normalized values.
[32]:
temp = filtered_prod_data[filtered_prod_data['randid'] == 'R10']
for xcol in ['irrad_poa_Wm2', 'temp_amb_C', 'wind_speed_ms']:
plt.scatter(temp[xcol], temp[prod_col_dict['capacity_normalized_power']])
plt.title(xcol)
plt.grid()
plt.show()



Dynamic linear modeling
We train a linear model using site data (irradiance, temperature, and windspeed)
[33]:
model_prod_data = filtered_prod_data.dropna(subset=['irrad_poa_Wm2', 'temp_amb_C', 'wind_speed_ms']+[prod_col_dict['powerprod']])
# Make sure to only pass data for one site! If sites are very similar, you can consider providing both sites.
model, train_df, test_df = linear.modeller(prod_col_dict,
kernel_type='polynomial',
#time_weighted='month',
X_parameters=['irrad_poa_Wm2', 'temp_amb_C'],#, 'wind_speed_ms'],
Y_parameter='generated_kW',
prod_df=model_prod_data,
test_split=0.05,
degree=3,
verbose=1)
train {1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12}
Design matrix shape: (8293, 108)
test {2, 3}
Design matrix shape: (437, 108)
Begin training.
[OLS] Mean squared error: 679448.77
[OLS] Coefficient of determination: 0.99
[OLS] 108 coefficient trained.
[RANSAC] Mean squared error: 812510.79
[RANSAC] Coefficient of determination: 0.98
Begin testing.
[OLS] Mean squared error: 7982573.16
[OLS] Coefficient of determination: 0.85
[OLS] 108 coefficient trained.
[RANSAC] Mean squared error: 81461712.61
[RANSAC] Coefficient of determination: -0.55
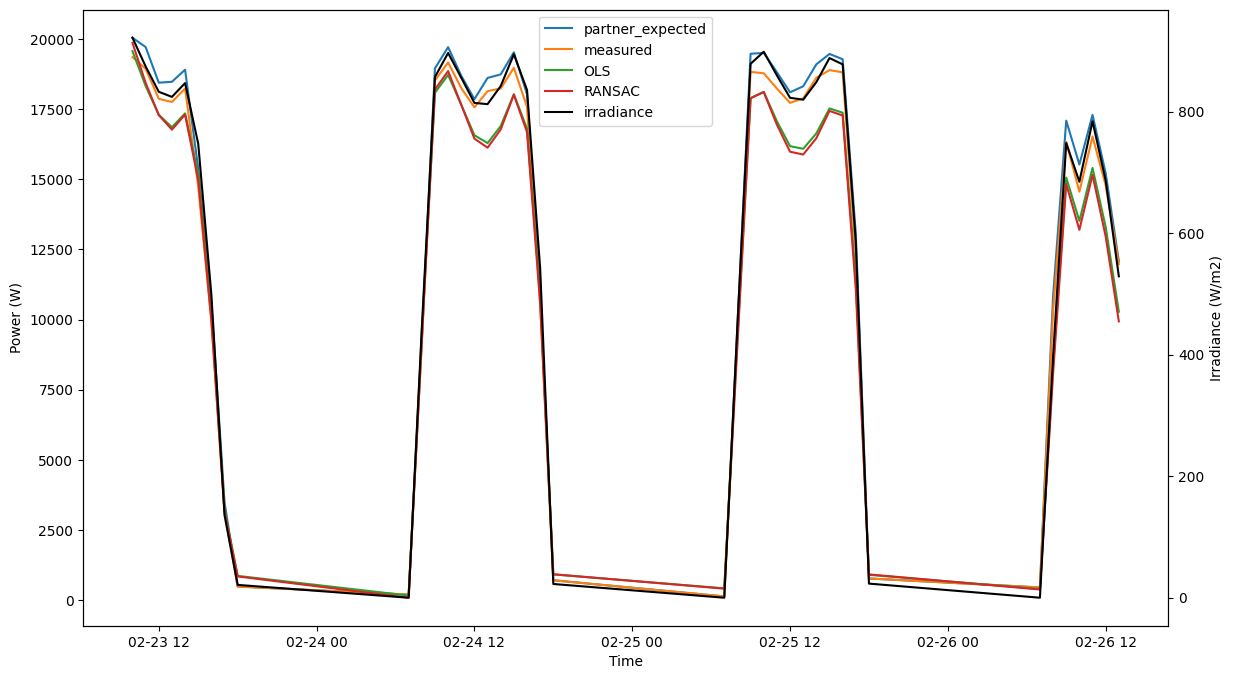
Define a plotting function
[34]:
from sklearn.metrics import mean_squared_error, r2_score
def plot(model, prod_col_dict, data_split='test', npts=50):
def print_info(real,pred,name):
mse = mean_squared_error(real, pred)
r2 = r2_score(real, pred)
print(f'[{name}] Mean squared error: %.2f'
% mse)
print(f'[{name}] Coefficient of determination: %.2f'
% r2)
fig,(ax) = plt.subplots(figsize=(14,8))
if data_split == 'test':
df = test_df
elif data_split == 'train':
df = train_df
measured = model.estimators['OLS'][f'{data_split}_y'][:npts]
ax2 = ax.twinx()
ax2.plot(model.estimators['OLS'][f'{data_split}_index'][:npts], df[prod_col_dict['irradiance']].values[:npts], 'k', label='irradiance')
ax.plot(model.estimators['OLS'][f'{data_split}_index'][:npts], df['expected_kW'].values[:npts], label='partner_expected')
print_info(measured, df['expected_kW'].values[:npts], 'partner_expected')
ax.plot(model.estimators['OLS'][f'{data_split}_index'][:npts], measured, label='measured')
for name, info in model.estimators.items():
predicted = model.estimators[name][f'{data_split}_prediction'][:npts]
ax.plot(model.estimators[name][f'{data_split}_index'][:npts], predicted, label=name)
print_info(measured, predicted, name)
ax2.set_ylabel("Irradiance (W/m2)")
ax.set_ylabel("Power (W)")
ax.set_xlabel('Time')
handles, labels = [(a+b) for a, b in zip(ax.get_legend_handles_labels(), ax2.get_legend_handles_labels())]
ax.legend(handles, labels, loc='best')
plt.show()
Observe performance
[35]:
plot(model, prod_col_dict, data_split='train', npts=40)
[partner_expected] Mean squared error: 2883252.79
[partner_expected] Coefficient of determination: 0.93
[OLS] Mean squared error: 225151.82
[OLS] Coefficient of determination: 0.99
[RANSAC] Mean squared error: 253701.63
[RANSAC] Coefficient of determination: 0.99

[36]:
plot(model, prod_col_dict, data_split='test', npts=40)
[partner_expected] Mean squared error: 273446.60
[partner_expected] Coefficient of determination: 0.99
[OLS] Mean squared error: 1037605.79
[OLS] Coefficient of determination: 0.98
[RANSAC] Mean squared error: 1300872.49
[RANSAC] Coefficient of determination: 0.97